-Search query
-Search result
Showing 1 - 50 of 317 items for (author: turner & hl)

EMDB-27920: 
3H03 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)
Method: single particle / : Turner HL, Ozorowski G, Ward AB

EMDB-27921: 
2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)
Method: single particle / : Turner HL, Ozorowski G, Ward AB

EMDB-27692: 
LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-27693: 
LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (global refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-8dt8: 
LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-27112: 
S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (global refinement)
Method: single particle / : Ozorowski G, Torres JL, Turner HL, Ward AB

EMDB-27113: 
S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement)
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-14784: 
AMC009 SOSIPv5.2 + ACS110 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14785: 
AMC009 SOSIPv.52 in complex with ACS114 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14786: 
AMC009 SOSIPv5.2 in complex with ACS117 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14787: 
AMC009 SOSIPv5.2 in complex with ACS122 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14788: 
AMC009 SOSIPv5.2 in complex with ACS125 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14789: 
AMC009 SOSIPv5.2 in complex with ACS131 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14783: 
AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124
Method: single particle / : van Schooten J, Ozorowski G, Ward A

EMDB-24693: 
SARS-CoV-2-6P-Mut7 S protein (C3 symmetry)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24694: 
SARS-CoV-2-6P-Mut7 S protein (asymmetric)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24695: 
CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24696: 
CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24697: 
CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24699: 
CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru1: 
SARS-CoV-2-6P-Mut7 S protein (C3 symmetry)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru2: 
SARS-CoV-2-6P-Mut7 S protein (asymmetric)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru3: 
CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru4: 
CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru5: 
CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru8: 
CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-14474: 
AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124
Method: single particle / : van Schooten J, Ward A

EMDB-14475: 
AMC009 SOSIPv5.2 + ACS101 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14476: 
AMC009 SOSIPv5.2 in complex with ACS102 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14477: 
AMC009 SOSIPv5.2 in complex with ACS103 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14478: 
AMC009 SOSIPv5.2 in complex with ACS124 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-23562: 
Negative stain EM map of 1E01 Fab in complex with N2 Singapore16
Method: single particle / : Turner HL, Ward AB

EMDB-23373: 
Negative stain EM map of RM20C Fab in complex with edc BG505 SOSIP.664
Method: single particle / : Turner HL, Ward AB, Cottrell CA
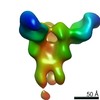
EMDB-23313: 
Negative stain map of monoclonal Fab 23 binding the lateral patch of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB

EMDB-23314: 
Negative stain map of monoclonal Fab 45 binding the esterase domain of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB
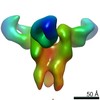
EMDB-23315: 
Negative stain map of monoclonal Fab 50 binding the lateral patch of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB
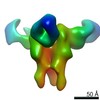
EMDB-23316: 
Negative stain map of monoclonal Fab 56 binding the lateral patch of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB

EMDB-23317: 
Negative stain map of monoclonal Fab 68 binding the Sa antigenic site of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB
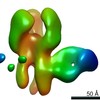
EMDB-23318: 
Negative stain map of monoclonal Fab 76 binding the stem of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB

EMDB-23319: 
CryoEM map of monoclonal Fabs 45 and 56 binding the esterase and lateral patch of H1 HA
Method: single particle / : Han J, Freyn AW, Turner HL, Ward AB
EMDB-22551: 
Polyclonal immune complex of Fab binding the receptor binding site region of H5 HA from serum of subject 36 at day 28
Method: single particle / : Ward A, Han J, Richey ST

EMDB-22552: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 36 at day 500
Method: single particle / : Ward A, Han J, Richey ST

EMDB-22553: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 7
Method: single particle / : Ward A, Han J, Richey ST

EMDB-22554: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 7
Method: single particle / : Ward A, Han J, Richey ST

EMDB-22555: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 21
Method: single particle / : Ward A, Han J, Richey ST

EMDB-22556: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 28
Method: single particle / : Ward A, Han J, Richey ST

EMDB-22557: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 28
Method: single particle / : Ward A, Han J, Richey ST
EMDB-22558: 
Polyclonal immune complex of Fab binding the receptor binding site region of H5 HA from serum of subject 43 at day 28
Method: single particle / : Ward A, Han J, Richey ST

EMDB-22559: 
Polyclonal immune complex of Fab binding the stem of H5 HA from serum of subject 43 at day 100
Method: single particle / : Ward A, Han J, Richey ST
EMDB-22560: 
Polyclonal immune complex of Fab binding the receptor binding site region of H5 HA from serum of subject 43 at day 100
Method: single particle / : Ward A, Han J, Richey ST
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
